The present study is a comprehensive SARS-CoV-2 genomic investigation performed in Amazonas, one of the Brazilian states most heavily impacted by COVID-19. Our analyses revealed that most cases in Amazonas were driven by successful dissemination of a few local viral clades that, together, comprise 77% of the 250 SARS-CoV-2 Amazonian genomes sampled between March 2020 and January 2021 rather than multiple importation events. Early major SARS-CoV-2 Amazonian clades arose in either Manaus or the metropolitan region between mid-March and late April 2020 and were widely disseminated within Amazonas state, reaching the most isolated inner localities. We found almost no evidence of the spread of early local SARS-CoV-2 Amazonian lineages outside the state, supporting that Amazonas was not a major hub of viral dissemination within Brazil during 2020. Increasing
travel during Christmas and New Year celebrations, combined with the emergence of a potentially more transmissible VOC, however, have changed this scenario, and lineage P.1 has rapidly spread across several Brazilian states up to March 2021 (http://www.genomahcov.fiocruz.br).
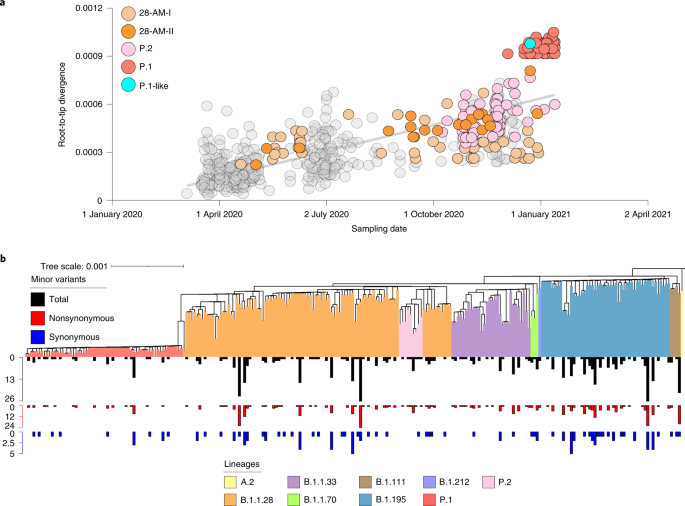
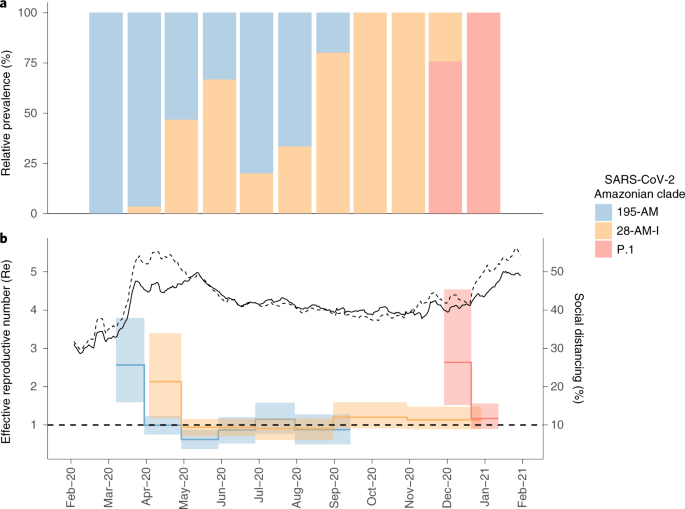
Two SARS-CoV-2 lineage replacements characterized the COVID-19 epidemic in Amazonas during early and late 2020. The first of these started after the primary epidemic peak and was a gradual process over nearly 5 months during which lineage B.1.1.28 progressively substitute this first lineage replacement. The SARS-CoV-2 Amazonian clades 28-AM-I and 28-AM-II, which became the dominant variants in the phase between peaks, displayed only single-lineage-defining synonymous mutations with one difference at the Spike protein when compared to clade 195-AM, and these evolved at a relatively constant rate between April and November 2020. The most notable difference was that clade 195-AM arose in the city of Manaus, and the Re value was considerably reduced around mid-April when social distancing in Manaus increased to >50%. Clades 28-AM-I and 28-AM-II, by contrast, arose outside the city of Manaus and the Re of clade 28-AM-I remained >1.0 until mid-May 2020, when social distancing outside the capital city increased to >50%. When mitigation measures were relaxed and the social distancing index fell by <40% in September 2020, the Re of clade 28-AM-I returned to >1.0 while clade B.1.195 became extinct, completing the lineage replacement process. Thus, the lower social distancing observed in Amazonas state inner municipalities compared to Manaus was the probable driver of the first lineaged replacement.
A modeling study with data from blood donors, conducted in Manaus, estimated that the first wave of SARS-CoV-2 infected 76% (95% CI: 67–98) of the city’s population by October 2020, suggesting that the theoretical threshold for herd immunity was reached by mid-2020 and that a second COVID-19 wave would not be expected so soon7. Several hypotheses were proposed to explain the unexpected second wave that resulted in the collapse of the health system in Manaus between December 2020 and January 2021 (ref. 5). One hypothesis is that lineage P.1 might evade immunity generated in response to a previous infection and has the potential to reinfect convalescent individuals. While some cases of reinfection with lineage P.1 were described in Manaus8, the extent to which reinfections effectively contribute to both onward transmission of SARS-CoV-2 and the surge of cases in the second wave in Amazonas remains controversial4,9. Another hypothesis is that the SARS-CoV-2 attack rate in Manaus was overestimated. Our study supports that a drastic reduction in median Re (from 2.1–2.6 to 0.9–1.0) for Amazonian lineages B.1.195 and B.1.1.28 occurred around April-May 2020, consistent with independent epidemiological modeling studies10,11, coinciding with the timing of implementation of nonpharmaceutical interventions (NPI) that effectively increased social distancing. Although those NPIs were not sufficiently stringent to consistently reduce Re to <1.0, they maintained a stationary state of low endemic viral community transmission from May to September 2020 (refs. 10,11). These findings support that the NPIs implemented in Amazonas brought the first epidemic wave under relative control before herd immunity became established, and were sufficiently effective to provide population ‘herd protection’ until December 2020 (ref. 11).
Mitigation measures were relaxed from September 2020 onwards, and the Re of clade 28-AM-I returned to >1.0. Nevertheless, the second epidemic wave started only in December 2020, coinciding with the emergence of the VOC P.1 and the second lineage replacement event. Several complementary evidences support that these events were probably driven by the emergence of a more transmissible variant in a context of relaxed social distancing. First, the second lineage replacement event was an abrupt process because the VOC P.1 emerged around late November 2020 and it required <2 months to become the dominant variant in Amazonas. This epidemiologic trajectory was later reproduced in other Brazilian states (http://www.genomahcov.fiocruz.br/). Second, the estimated median Re of the VOC P.1 during December 2020 was 2.2-fold higher than that estimated for clade 28-AM-I in the same period, indicating that P.1 could have been nearly twofold more transmissible than the co-circulating B.1.1.28 parental lineage, consistent with two recent independent studies4,9.
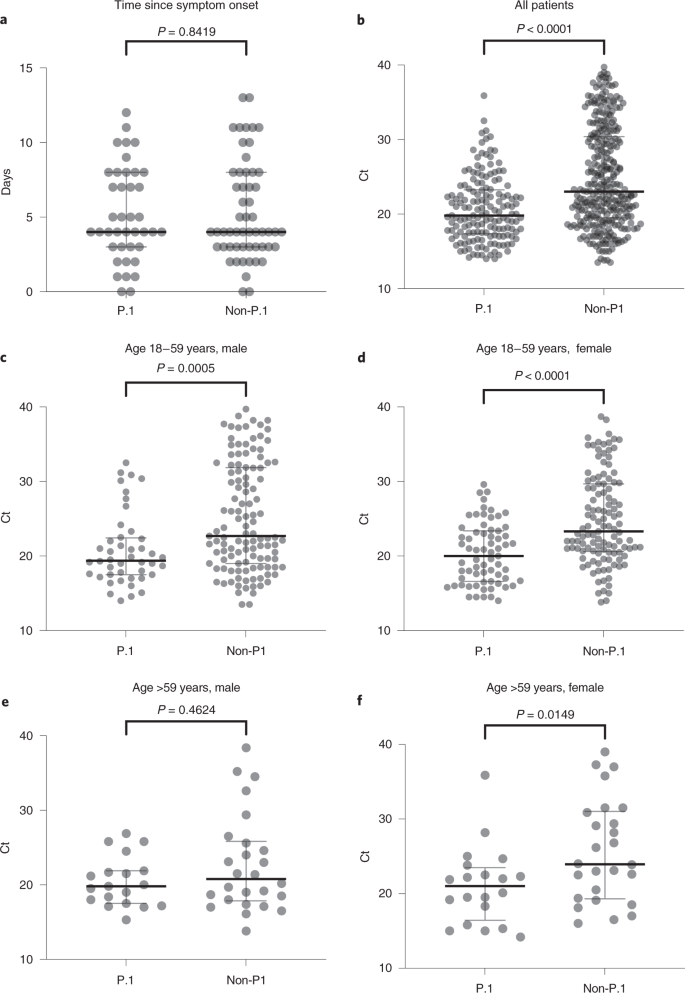
Third, the level of SARS-CoV-2 RNA (estimated from the median Ct) in the URT samples from P.1 infections, particularly from adults (18–59 years), was ~tenfold higher than the level detected in non-P.1 infections, suggesting that P.1-infected adult individuals are more infectious than those harboring non-P.1 viruses4,9. Fourth, recent experimental evidence supports that VOC P.1 displayed both higher affinity for the human receptor ACE2 and increased resistance to antibody neutralization6,12,13,14, which might provide a substantial selective advantage for transmission of P.1 over other lineages.
Understanding the factors that drive the emergence and expansion of VOCs harboring multiple key mutations in the receptor-binding domain of the Spike protein is of crucial importance. One hypothesis is that the emergence of VOCs resulted from a major change in the selective environment, probably imposed by partial herd immunity in heavily affected regions within which SARS-CoV-2 was evolving15. Our study of the Amazonian clades that locally evolved between April and December 2020, however, revealed no unusual patterns of intra- or interhost viral variability, showing that the local emergence of VOC is an
2021 (ref. 17)—of P.1-like viruses that harbor several of the P.1 lineage-defining mutations indicates that P.1 mutations did not accumulate in a single, long-term infection but were acquired in sequential steps as observed in the VOC B.1.351 (ref. 18). The finding that P.1 and P.1-like viruses probably share a most recent common ancestor in late August 2020 further supports that SARS-CoV-2 variants carrying mutations of concern had circulated in Manaus for some time before the emergence of lineage P.1. Although only the lineage P.1 seems to have displayed a rapid dissemination to date, our findings alert for the potential spread of other P.1-related VOC in Amazonas state, or in other Brazilian states.
It is important to stress that our study has some limitations. First, biased sampling across Brazilian states might influence phylogeographic reconstructions of within-country B.1.128 and B.1.1.33 lineage migrations, so the inferred number of importations into Amazonas should be interpreted as lower-bound estimates. Second, within-country spread of lineages B.1.1.28 and B.1.1.33 was inferred using a machine learning phylogeographic approach that does not account for uncertainty in phylogenetic reconstruction, and thus the routes of viral migration described here are plausible hypotheses among alternatives not fully explored in our analyses. Third, estimates of Re obtained here may be influenced by local epidemiological dynamics, making it challenging to extrapolate the difference in viral transmissibility between P.1 and non-P.1 variants observed in Amazonas to other geographic
regions. Fourth, although we removed potential confounders (for example, comparison of only PCR results using the same RNA extraction/real-time RT–PCR protocols and sampled at similar timepoints from symptom onset), Ct comparisons have weaknesses and must be analyzed with caution. VOC P.1 may cause more prolonged infections with a similar peak in viral concentration than non-P.1 lineages, as was recently described for B.1.1.7 (ref. 19). Moreover, we have no data regarding disease severity for group comparison. Therefore, the difference observed here should be confirmed in other geographic settings, including analysis of Ct dynamics in longitudinal sampled patients with different disease outcomes.
In summary, our findings support that lineage replacements were a recurrent phenomenon in the local evolution of SARS-CoV-2 in Amazonas state, driven by ecological and virological factors. Our findings also indicate that NPIs deployed in Amazonas state in April 2020 were sufficiently effective to reduce the Re of early prevalent local SARS-CoV-2 clades but were insufficient to keep the epidemic under control, allowing the establishment and local persistence of several endemic viral lineages and subsequent emergence of the VOC P.1 in late November/early December 2020. The lack of efficient social distancing and other mitigation measures probably allowed a sudden and accelerated transmission of VOC P.1. At the same time, the higher transmissibility of this VOC further fueled the rapid upsurge in SARS-CoV-2 cases and hospitalizations observed in Manaus following its emergence. Importantly, phylodynamic modeling indicates that NPIs implemented in Manaus since early January 2021 (Supplementary Note) effectively reduced the median Re of the VOC P.1 by approximately 50%. Therefore, our results suggest that weak adoption of NPIs represents a risk for the continuous emergence of new variants. Implementation of efficient mitigation measures, combined with widespread vaccination, will be crucial to controlling the spread of SARS-CoV-2 VOCs in Brazil.
Methods
SARS-CoV-2 samples and ethical aspects
We collected nasopharyngeal and pharyngeal swabs from 644 residents in Amazonas state with available demographic data (320 male, median age 44 years (interquartile range (IQR) = 31.0–57.7); 324 female, median age 43 years (IQR = 30.2–56.0)) that were positively tested by real-time RT–PCR as a routine diagnostic for COVID-19 using any of the following commercial assays: SARS-CoV2 (E/RP) (Biomanguinhos), Allplex 2019-nCoV Assay (Seegene) or an in-house protocol following US Centers for Disease Control and Prevention (CDC) guidelines (https://www.fda.gov/media/134922/download). Among those 644
nasopharyngeal and pharyngeal swab samples, 250 were submitted to nucleotide sequencing and 394 were evaluated only for P.1/VOCs by the real-time RT–PCR developed in this study. Fiocruz/ILMD is one of the official laboratories designated for SARS-CoV-2 testing under the auspices of a network coordinated by the Amazonas State Health Foundation (FVS-AM) and the Brazilian Ministry of Health. This study was conducted at the request of the SARS-CoV-2 surveillance program of FVS-AM. It was approved by the Ethics Committee of Amazonas State University (no. 25430719.6.0000.5016), which waived signed informed consent.
Detection of SARS-CoV-2 P.1/VOCs by RT–PCR
A total of 1,626 SARS-CoV-2-positive samples collected between 1 November 2020 and 31 January 2021 (including those 394 with demographic data) were submitted to a real-time RT–PCR screening test designed for the detection of VOCs that use a forward primer (P.1/VOCs-FNF 5′- GGGTGATGCGTATTATGACATGGTTGG), a reverse primer (P.1/VOCs-FNR 5′- CTAGCACCATCATCATACACAGTTCTTGC) and a probe (P.1/VOCs-FNP 5′ FAM (ZEN)- TGGTTGATACTAGTTTGAAGCTAAAA), to detect the ORF1b deletion (NSP6: S106del, G107del, F108del) found in the three VOCs (P.1, B.1.1.7 and B.1.351). Both primers were used at 300 nM and the probe at 150 nM (final concentration), with TaqMan one-step Fast Virus master Mix (ThermoFisher Scientific, no. 4444434). All real-time RT–PCR data collected in this experiment were acquired using the QuantStudio 5 Real-Time PCR System and QuantStudio design & analysis software v.1.4.1 (ThermoFisher Scientific). We previously validated this assay against 185 high-quality, full SARS-CoV-2 genomes, 59 non-P.1 and 126 P.1
(Supplementary Table 7). All oligos used in this study were manufactured by IDT DNA. Because we have not detected the circulation of VOCs B.1.1.7 and B.1.351 in Amazonas state, we use the frequency of NSP6 deletion among real-time RT–PCR positives as a reliable proxy for frequency of the VOC P.1.
SARS-CoV-2 amplification and sequencing
A total of 250 SARS-CoV-2-positive samples (122 male, 128 female; median age 43 years, IQR = 32-46) collected from residents of 25 out of 62 municipalities in Amazonas state, including the capital Manaus, between 16 March and 13 January were subjected to amplification and next-generation sequencing as previously described1, now with a reduced number of amplicons (nine rather than 15) of mean average size ~3,500 bp (ref. 8). Briefly, RNAs were extracted with the Maxwell RSC Viral Total Nucleic Acid Purification Kit (Promega, no. AS1330) and then converted to complementary DNA with Superscript IV reverse transcriptase (ThermoFisher Scientific, no. 18090200). Amplicons were amplified with SuperFi II Green PCR master mix (a proofreading DNA polymerase with >300× Taq fidelity from ThermoFisher Scientific (no. 12369010)), precipitated with PEG 8000 (Promega, no. V3011) and quantified using a fluorimeter. Normalized pooled amplicons of each sample
were used to prepare next-generation sequencing libraries with Nextera XT (no. FC-131-1096) and clustered with 500 cycles of MiSeq Reagent Kit v.2 (no. MS-102-2003) on 2× 250 cycles or 2× 150 cycles (no. MS-103-1002) of paired-end runs. All sequencing data were collected using the MiSeq sequencing platform and Miseq Control software v.2.6.2.1 (Illumina).
SARS-CoV-2 whole-genome consensus sequences and genotyping
FASTQ reads were generated by the Illumina pipeline at BaseSpace (https://basespace.illumina.com). All files were downloaded and imported into Geneious v.10.2.6 for trimming and assembly using a customized workflow employing BBDuk and BBMap tools (v.37.25) and the NC_045512.2 RefSeq as a template. Using a threshold of at least 50% to call a base, we generated consensus sequences with mean depth coverage of 2,689× (95% CI of mean 2,376–3,002), with only eight genomes having <1,000× depth coverage. The mean number of mapped reads was 512,967 (95% CI of mean 441,779–584,154), covering at least 98.9% of the RefSeq genome. The final consensus sequences had at least 94% bases with Q score = 30, zero ambiguities and were carefully inspected when a disagreement with RefSeq was observed. Coverage and pairwise identity percentages, as well as the total number of mapped reads (without duplicates) to RefSeq and the percentage of high-quality bases in consensus, were calculated for all consensus files and are shown in Supplementary Table 1. Consensus sequences were initially assigned to viral lineages according to the nomenclature proposed by Rambaut et al.20, using the Pangolin web application (https://pangolin.cog-uk.io) and later confirmed by phylogenetic analyses.
Intrahost SARS-CoV-2 genomic variability
Raw sequencing reads and primer sequences were removed with Trimmomatic v.0.26 (ref. 21) using default parameters. Reads that passed quality filtering were then mapped against the reference genome (NC_045512.2) using the Bowtie2 software v.2.3.5.1 (ref. 22). A.bed file was generated with bedtools v.2.15.0 (ref. 23), SAMtools v.1.10 (ref. 24) and vcftools v.0.1.13 (ref. 25) using the internal parameter vcf-annotated (parameters-filter Qual = 20/MinDP = 100/SnpGap = 20), meaning that only those nucleotide variants supported by reads with mapping quality >20 and at least 100 sequencing coverage depth would be retained in the intermediate variant call file. To characterize the viral intrahost population, we identified all MVs found in the samples—that is, nucleotides highly supported by 10–49% of the reads in a given position and that were not included in the final majority consensus genome. We then replaced the nucleotides supported by the majority of reads by MVs in the consensus genome to evaluate the impact of synonymous and nonsynonymous nucleotide variation between major and minor variants. We performed the synonymous and nonsynonymous analysis using an R pipeline developed for SARS-CoV-2 (ref. 26) with R v.4.0.3 and RStudio
v.1.4.1103.
Discrete maximum likelihood and Bayesian phylogeography
All high-quality (<1% N, or non-identified nucleotide) complete (>29 kb) SARS-CoV-2 genomes of lineages B.1.1.28 (n = 512) and B.1.1.33 (n = 595) sampled in Brazil, and of lineage B.1.195 sampled worldwide (n = 110), that were available on GISAID (https://www.gisaid.org/) as of 13 January 2021, were downloaded. SARS-CoV-2 complete genome sequences were aligned using MAFFT v.7.475 (ref. 27). The B.1.1.28 and B.1.1.33 datasets were subjected to maximum likelihood phylogenetic analysis using IQ-TREE v.2.1.2 (ref. 28) under a general time-reversible (GTR) model of nucleotide substitution with a gamma-distributed rate variation among sites and four rate categories (G4), a proportion of invariable sites (+I) and empirical base frequencies (+F), as selected by the ModelFinder application29,30. Branch support was assessed by the approximate likelihood-ratio test based on the Shimodaira–Hasegawa-like procedure with 1,000 replicates. Time-scaled phylogeographic maximum likelihood phylogenetic trees of Brazilian B.1.1.28 and B.1.1.33 datasets were reconstructed using Treetime v.0.8.1 (ref. 31), with a fixed substitution rate (8 × 10–4 substitutions per site per year) coupled with an ancestral character reconstruction of epidemic locations using PASTML v.1.9.15 (ref. 32) with marginal posterior probabilities approximation and an F81-like model. A time-scaled Bayesian phylogeographic analysis was performed for B.1.195 sampled worldwide using the Bayesian Markov chain Monte Carlo (MCMC) approach, implemented in BEAST v.1.10.4 (ref. 33) with BEAGLE library v.3 (ref. 34), to improve computational time. The Bayesian tree was reconstructed using the GTR + F + I + G4 nucleotide substitution model, the nonparametric Bayesian skyline model as the coalescent tree prior35, a strict
molecular clock model with a uniform substitution rate prior (8–10 × 10-4 substitutions per site per year) and a reversible discrete phylogeographic model36 with a continuous-time Markov chain (CTMC) rate reference prior37. Additionally, the nine-nucleotide deletion at nsp1 (delta 640–648: K141, S142, F143) characteristic of the 195-AM clade was incorporated as an informative trait in phylogenetic reconstruction, and transitions were modeled with a symmetric CTMC rate prior. Three MCMC chains were run for 100 million generations and then combined to ensure stationarity and good mixing. Convergence (effective sample size >200) in parameter estimates was assessed using TRACER v.1.7 (ref. 38). The maximum clade credibility (MCC) tree was summarized with TreeAnnotator v.1.10. Maximum likelihood and MCC trees were visualized using FigTree v.1.4.4 (http://tree.bio.ed.ac.uk/software/figtree/).
Continuous Bayesian phylogeography
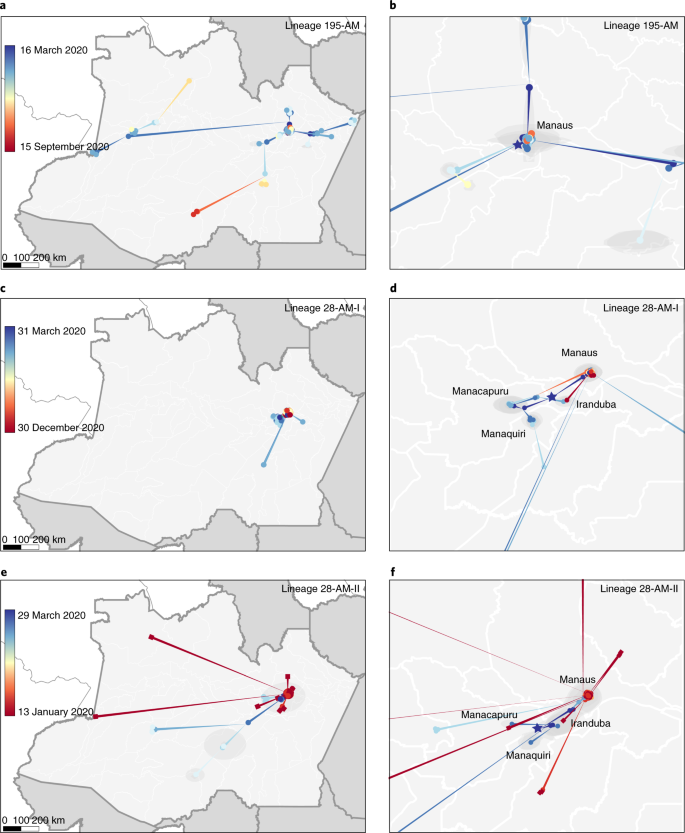
The phylogenetic diffusion of SARS-CoV-2 clades from Amazonas state identified by the maximum likelihood analysis (195-AM, 28-AM-I and 28-AM-II) was estimated with the heterogeneous relaxed random walk model and a Cauchy distribution39, previously applied to SARS-CoV-2 in Brazil40, using BEAST v.1.10.4 (ref. 33) as explained above. We used strict and local molecular clock models41,42 with a uniform substitution rate prior (8–10 × 10–4 substitutions per site per year) to estimate evolutionary rates. Viral spatial–temporal diffusion was analyzed and visualized in SPREAD v.1.0.7 (ref. 43), and further projected in maps generated with QGIS v.3.10.2 software (http://qgis.org) using public access data downloaded from the GADM v.3.6 database (https://gadm.org). For each lineage and molecular clock model, one MCMC chain was run for 150 million generations and
stationarity and mixing were checked as explained above.
Data for social distancing trends
The social distancing trends were obtained from a commercial company (http://inloco.com.br). Inloco’s isolation index analyzes people’s movements for different levels, states, cities and microregions inferred from proprietary technology. Thus, the higher the index the greater the degree of isolation estimated from the site. This index has been used by several Brazilian states’ decision-making authorities since the beginning of the pandemic.
Estimation of Re
To estimate the Re of the Amazonian SARS-CoV-2 clades over time we used the BDSKY model44 implemented within BEAST 2 v.2.6.2 (ref. 45). The sampling rate (d) was set to zero for the period before the oldest sample and then estimated from the data. The BDSKY prior settings were as follows: become uninfectious rate (exponential, mean = 36); reproductive number (log normal, mean = 0.8, s.d. = 0.5); sampling proportion (beta, alpha = 1, beta = 100). Origin parameter was conditioned to root height, and Re was estimated in a piecewise manner over six time intervals (monthly) to the 195-AM clade, five time intervals (bimonthly) to the 28-AM-I clade and two equal time intervals to the P.1 clade. Time intervals were
defined from the date of the most recent sample up to the root of the tree. The molecular clock and substitution model were as in the phylogeographic analysis. One MCMC chain was run for 20 million generations and then checked for stationarity and mixing, as explained above.
Statistical analysis
Descriptive statistics, testing for normal distribution (D’Agostino and Pearson and Anderson–Darling) and the nonparametric Mann–Whitney test were used to compare the Ct of SARS-CoV-2 RT–PCR-positive samples from the URT of patients infected with P.1 versus non-P.1 viruses. To avoid bias, only Ct values from samples analyzed by the same RNA
extraction method (Promega Maxwell) and the same real-time RT–PCR diagnostic assay (for example, the CDC assay) were compared. The threshold for statistical significance was set to P < 0.05 using two-sided tests. Graphics and statistical analyses were performed using GraphPad v.9.01 and v.9.02 (Prism Software).
Reporting Summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Data availability
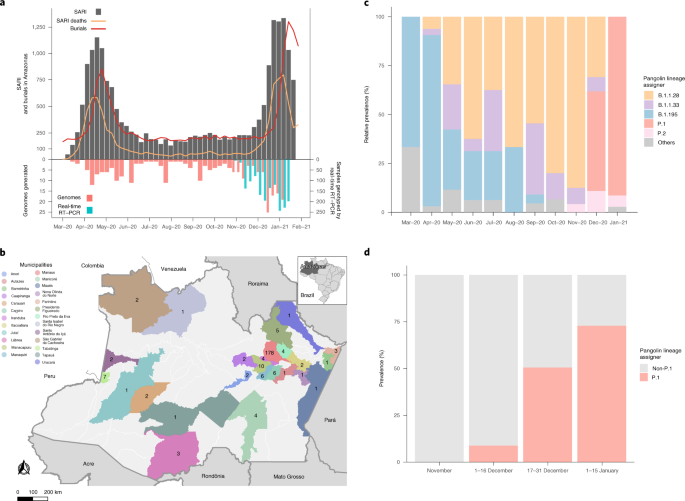
All SARS-CoV-2 genomes generated and analyzed in this study are available at the EpiCoV database in GISAID (https://www.gisaid.org) under IDs EPI_ISL_792560, EPI_ISL_801386–801403, EPI_ISL_811148, EPI_ISL_811149, EPI_ISL_833131–833140, EPI_ISL_1034304–1034306, EPI_ISL_1068078–1068292 and EPI_ISL_1661250–1661252. Figures 1a and 5b were created with data provided by http://info.gripe.fiocruz.br, SEMULSP-Manaus and FVS-AM. Administrative areas presented in Fig. 4 were provided by the GADM v.3.6 database (http://gadm.org). Detailed results of MV detection are available at GitHub (https://github.com/dezordi/mFinder/tree/naveca_et_al_2021/supplementary_data). Source
data are provided with this paper.
References
- 1.
Nascimento, V. A. D. et al. Genomic and phylogenetic characterisation of an imported case of SARS-CoV-2 in Amazonas State, Brazil. Mem. Inst. Oswaldo Cruz 115, e200310 (2020).
CAS Article Google Scholar
- 2.
Fundação em Vigilância e Saúde do Amazonas. Boletim diários dos casos de COVID-19. https://www.fvs.am.gov.br/media/publicacao/21_02_21_BOLETIM_DIÁRIO_DE_CASOS_COVID-19.pdf (2021).
- 3.
Fujino, T. et al. Novel SARS-CoV-2 variant identified in travelers from Brazil to Japan. Emerg. Infect. Dis. https://doi.org/10.3201/eid2704.210138 (2021).
- 4.
Faria, N. R. et al. Genomics and epidemiology of the P.1 SARS-CoV-2 lineage in Manaus, Brazil. Science https://doi.org/10.1126/science.abh2644 (2021).
- 5.
Sabino, E. C. et al. Resurgence of COVID-19 in Manaus, Brazil, despite high seroprevalence. Lancet 397, 452–455 (2021).
CAS Article Google Scholar
- 6.
Dejnirattisai W. et al. Antibody evasion by the P.1 strain of SARS-CoV-2. Cell https://doi.org/10.1016/j.cell.2021.03.055 (2021).
- 7.
Buss, L. F. et al. Three-quarters attack rate of SARS-CoV-2 in the Brazilian Amazon during a largely unmitigated epidemic. Science 371, 288–292 (2021).
- 8.
Naveca, F. et al. Three SARS-CoV-2 reinfection cases by the new Variant of Concern (VOC) P.1/501Y.V3. Preprint at Res. Sq. https://doi.org/10.21203/rs.3.rs-318392/v1 (2021).
- 9.
Coutinho, R. M. et al. Model-based estimation of transmissibility and reinfection of SARS-CoV-2 P.1 variant. Preprint at medRxiv https://doi.org/10.1101/2021.03.03.21252706 (2021).
- 10.
Mellan, T. A. et al. Subnational analysis of the COVID-19 epidemic in Brazil. Preprint at medRxiv https://doi.org/10.1101/2020.05.09.20096701 (2020).
- 11.
He, D., Artzy-Randrup, Y., Musa, S. S. & Stone, L. The unexpected dynamics of COVID-19 in Manaus, Brazil: was herd immunity achieved? Preprint at medRxiv https://doi.org/10.1101/2021.02.18.21251809 (2021).
- 12.
Hoffmann, M. et al. SARS-CoV-2 variants B.1.351 and P.1 escape from neutralizing antibodies. Cell https://doi.org/10.1016/j.cell.2021.03.036 (2021).
- 13.
Garcia-Beltran, W. F. et al. Multiple SARS-CoV-2 variants escape neutralization by vaccine-induced humoral immunity. Cell https://doi.org/10.1016/j.cell.2021.03.013 (2021).
- 14.
Wang, P. et al. Increased resistance of SARS-CoV-2 variant P.1 to antibody neutralization.
-
Cell Host Microbe https://doi.org/10.1016/j.chom.2021.04.007 (2021).
- 15.
Martin, D. P. et al. The emergence and ongoing convergent evolution of the N501Y lineages coincides with a major global shift in the SARS-CoV-2 selective landscape. Preprint at medRxiv https://doi.org/10.1101/2021.02.23.21252268 (2021).
- 16.
McCormick, K. D., Jacobs, J. L. & Mellors, J. W. The emerging plasticity of SARS-CoV-2. Science 371, 1306–1308 (2021).
- 17.
Resende, P. C. et al. The ongoing evolution of variants of concern and interest of SARS-CoV-2 in Brazil revealed by convergent indels in the amino (N)-terminal domain of the Spike protein. Preprint at medRxiv https://doi.org/10.1101/2021.03.19.21253946 (2021).
- 18.
Tegally, H. et al. Detection of a SARS-CoV-2 variant of concern in South Africa. Nature https://doi.org/10.1038/s41586-021-03402-9 (2021).
- 19.
Kissler, S. M. et al. Densely sampled viral trajectories suggest longer duration of acute infection with B.1.1.7 variant relative to non-B.1.1.7 SARS-CoV-2. Preprint at medRxiv https://doi.org/10.1101/2021.02.16.21251535 (2021).
- 20.
Rambaut, A. et al. A dynamic nomenclature proposal for SARS-CoV-2 lineages to assist genomic epidemiology. Nat. Microbiol. https://doi.org/10.1038/s41564-020-0770-5 (2020).
- 21.
Bolger, A. M., Lohse, M. & Usadel, B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014).
CAS Article Google Scholar
- 22.
-
Langmead, B. & Salzberg, S. L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012).
CAS Article Google Scholar
- 23.
Quinlan, A. R. BEDTools: the Swiss-Army tool for genome feature analysis. Curr. Protoc. Bioinformatics 47, 11.12.1–34 (2014).
Google Scholar
- 24.
Li, H. et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079 (2009).
- 25.
Danecek, P. et al. The variant call format and VCFtools. Bioinformatics 27, 2156–2158 (2011).
CAS Article Google Scholar
- 26.
Mercatelli, D. & Giorgi, F. M. Geographic and genomic distribution of SARS-CoV-2 mutations. Front. Microbiol. 11, 1800 (2020).
Article Google Scholar
- 27.
Katoh, K. & Standley, D. M. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780 (2013).
- 28.
Nguyen, L. T., Schmidt, H. A., von Haeseler, A. & Minh, B. Q. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32, 268–274 (2015).
CAS Article Google Scholar
- 29.
Tavaré, S. Some probabilistic and statistical problems in the analysis of DNA sequences. Lectures on Mathematics in the Life Sciences
-
https://www.damtp.cam.ac.uk/user/st321/CV_&_Publications_files/STpapers-pdf/T86.pdf (1986).
- 30.
Kalyaanamoorthy, S., Minh, B. Q., Wong, T. K. F., von Haeseler, A. & Jermiin, L. S. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat. Methods 14, 587–589 (2017).
- 31.
Sagulenko, P., Puller, V. & Neher, R. A. TreeTime: maximum-likelihood phylodynamic analysis. Virus Evol. https://doi.org/10.1093/ve/vex042 (2018).
- 32.
Ishikawa, S. A., Zhukova, A., Iwasaki, W. & Gascuel, O. A fast likelihood method to reconstruct and visualize ancestral scenarios. Mol. Biol. Evol. 36, 2069–2085 (2019).
CAS Article Google Scholar
- 33.
Suchard, M. A. et al. Bayesian phylogenetic and phylodynamic data integration using BEAST 1.10. Virus Evol. 4, vey016 (2018).
Article Google Scholar
-
- 34.
Suchard, M. A. & Rambaut, A. Many-core algorithms for statistical phylogenetics. Bioinformatics 25, 1370–1376 (2009).
CAS Article Google Scholar
- 35.
Drummond, A. J., Rambaut, A., Shapiro, B. & Pybus, O. G. Bayesian coalescent inference of past population dynamics from molecular sequences. Mol. Biol. Evol. 22, 1185–1192 (2005).
CAS Article Google Scholar
- 36.
Lemey, P., Rambaut, A., Drummond, A. J. & Suchard, M. A. Bayesian phylogeography finds its roots. PLoS Comput. Biol. 5, e1000520 (2009).
Article Google Scholar
- 37.
Ferreira, M. A. R. & Suchard, M. A. Bayesian analysis of elapsed times in continuous‐time Markov chains. Can. J. Stat. 36, 355–368 (2008).
Article Google Scholar
- 38.
Rambaut, A., Drummond, A. J., Xie, D., Baele, G. & Suchard, M. A. Posterior summarisation in Bayesian phylogenetics using Tracer 1.7. Syst. Biol. https://doi.org/10.1093/sysbio/syy032 (2018).
- 39.
Lemey, P., Rambaut, A., Welch, J. J. & Suchard, M. A. Phylogeography takes a relaxed random walk in continuous space and time. Mol. Biol. Evol. 27, 1877–1885 (2010).
CAS Article Google Scholar
- 40.
Candido, D. S. et al. Evolution and epidemic spread of SARS-CoV-2 in Brazil. Science 369, 1255–1260 (2020).
CAS Article Google Scholar
- 41.
Drummond, A. J. & Suchard, M. A. Bayesian random local clocks, or one rate to rule them all. BMC Biol. 8, 114 (2010).
Article Google Scholar
- 42.
Yoder, A. D. & Yang, Z. Estimation of primate speciation dates using local molecular clocks. Mol. Biol. Evol. 17, 1081–1090 (2000).
CAS Article Google Scholar
- 43.
Bielejec, F., Rambaut, A., Suchard, M. A. & Lemey, P. SPREAD: spatial phylogenetic reconstruction of evolutionary dynamics. Bioinformatics 27, 2910–2912 (2011).
CAS Article Google Scholar
- 44.
Stadler, T., Kuhnert, D., Bonhoeffer, S. & Drummond, A. J. Birth–death skyline plot reveals temporal changes of epidemic spread in HIV and hepatitis C virus (HCV). Proc. Natl Acad. Sci. USA 110, 228–233 (2013).
CAS Article Google Scholar
- 45.
Bouckaert, R. et al. BEAST 2.5: an advanced software platform for Bayesian evolutionary analysis. PLoS Comput. Biol. 15, e1006650 (2019).
Acknowledgements
We thank all the health care workers and scientists who have worked hard to deal with this pandemic threat, the GISAID team and all the EpiCoV database submitters—in particular, the Japanese National Institute of Infectious Diseases members T. Sekizuka, K. Itokawa, R. Tanaka and M. Hashino—for publication of genomes. A GISAID acknowledgment table containing sequences used in this study is shown in Supplementary Table 8. We also thank N. Faria for sharing unpublished findings regarding the SARS-CoV-2 B.1.1.28 lineage. We also appreciate the support of Genomic Coronavirus Fiocruz Network members and the Respiratory Viruses Genomic Surveillance Network of the General Laboratory Coordination of the Brazilian Ministry of Health, Brazilian Central Laboratory States and the Amazonas surveillance teams for their partnership in viral surveillance in Brazil. Funding support is
acknowledged from Fundação de Amparo à Pesquisa do Estado do Amazonas – FAPEAM (PCTI-EmergeSaude/AM call 005/2020 and Rede Genômica de Vigilância em Saúde), Conselho Nacional de Desenvolvimento Científico e Tecnológico – CNPq (grant no. 403276/2020-9) and Inova Fiocruz/Fundação Oswaldo Cruz (grant no. VPPCB-007-FIO-18-2-30—Geração de conhecimento), received by F.G.N., and Fundação de Amparo à Pesquisa do Estado do Rio de Janeiro – FAPERJ (grant no. E-26/202.896/2018), received by G.B. F.G.N. and G.B. are also supported by CNPq through their productivity research fellowships (306146/2017-7 and 302317/2017-1, respectively). In loving memory of F. R. Naveca, C. R. dos Santos, V. L. Costa de Souza and all the relatives and colleagues we have lost to COVID-19.
Author information
Affiliations
-
Laboratório de Ecologia de Doenças Transmissíveis na Amazônia, Instituto Leônidas e Maria Deane, Fiocruz, Manaus, Brazil
Felipe Gomes Naveca, Valdinete Nascimento, Victor Costa de Souza, André de Lima Corado, Fernanda Nascimento, George Silva, Ágatha Costa, Débora Duarte, Karina Pessoa, Matilde Mejía & Maria Júlia Brandão
-
Laboratório de Diversidade Microbiana da Amazônia com Importância para a Saúde,
-
Instituto Leônidas e Maria Deane, Fiocruz, Manaus, Brazil
Michele Jesus
-
Fundação de Vigilância em Saúde do Amazonas, Manaus, Brazil
Luciana Gonçalves, Cristiano Fernandes da Costa, Vanderson Sampaio & Daniel Barros
-
Laboratório Central de Saúde Pública do Amazonas, Manaus, Brazil
Marineide Silva & Tirza Mattos
-
Instituto Nacional de Pesquisas da Amazônia, Manaus, Brazil
Gemilson Pontes
-
Universidade do Estado do Amazonas, Manaus, Brazil
Ligia Abdalla
-
Hospital Adventista de Manaus, Manaus, Brazil
João Hugo Santos
-
Laboratório de AIDS e Imunologia Molecular, Instituto Oswaldo Cruz, Fiocruz, Rio de Janeiro, Brazil
Ighor Arantes & Gonzalo Bello
-
Instituto Aggeu Magalhães, Departamento de Entomologia e Núcleo de Bioinformática, Fiocruz, Recife, Brazil
-
Filipe Zimmer Dezordi & Gabriel Luz Wallau
-
Laboratório de Vírus Respiratórios e Sarampo, Instituto Oswaldo Cruz, Fiocruz, Rio de Janeiro, Brazil
Marilda Mendonça Siqueira & Paola Cristina Resende
-
Departamento de Biologia, Centro de Ciências Exatas, Naturais e da Saúde, Universidade Federal do Espírito Santo, Alegre, Brazil
Edson Delatorre
-
Instituto Gonçalo Moniz, Fiocruz, Salvador, Brazil
-
Tiago Gräf
Contributions
F.G.N. contributed to writing of the report, data analysis, laboratory management and obtaining financial support. V.N., V.C.d.S., A.d.L.C., F.N., G.S., A.C., D.D., K.P., M.M., M.J.B., M.J. and L.G. contributed to diagnostics and sequencing analysis. C.F.d.C., V.S., D.B., M.S., T.M., G.P., L.A. and J.H.S. contributed to patient and public health surveillance data. I.A. and F.Z.D. contributed to formal data analysis of sequence diversity. M.M.S., G.L.W., P.C.R., E.D., T.G. and G.B. contributed to formal data analysis and writing and editing of the report.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature Medicine thanks Richard Neher, Xiang Ji and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Alison Farrell
was the primary editor on this article and managed its editorial process and peer review in collaboration with the rest of the editorial team.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
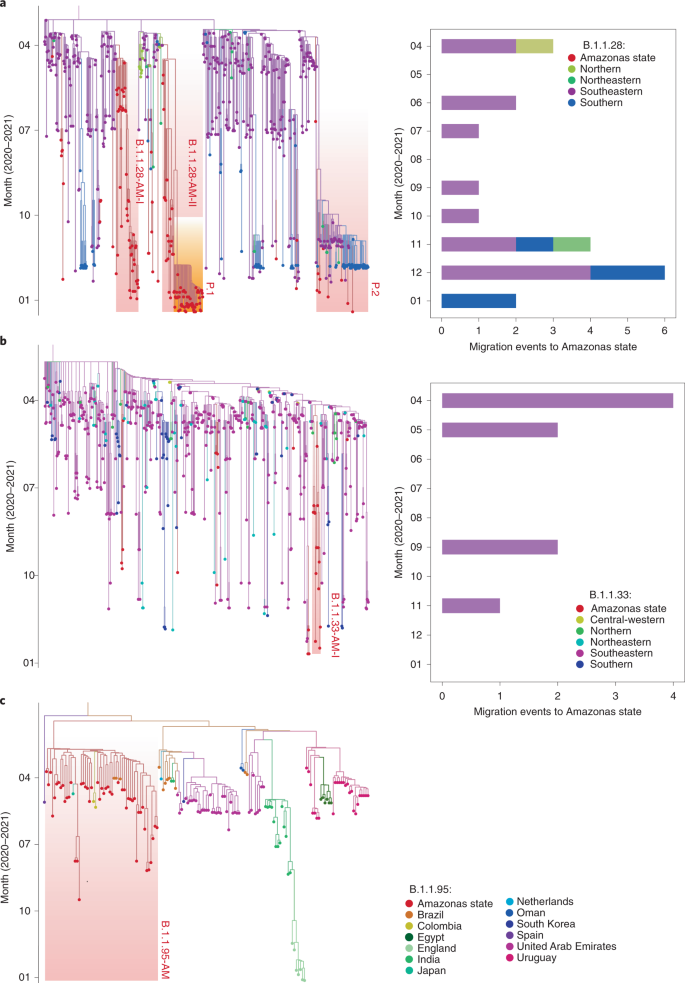
ML phylogeographic analysis of lineages B.1.1.28/P.2 (n = 674) (a) and B.1.1.33 (n = 602) (b) in Brazil. Ancestral character state reconstruction was done in PastML with time-scaled trees. Importation and exportation events deduced from location state changes toward (n = 28) and from (n = 2) the Amazon state are detailed. The singleton Amazonian sequences and the MRCA of Amazonian clusters are indicated by black outlined circular shapes. Shaded boxes indicate the Amazonian clusters and the sub-clades that define lineages P.1 and P.2. The origin or destination of the event is indicated alongside the estimated date (Amazonian clusters) or sampling date (singletons). All locations are colored according to the legend in the bottom right.
Violin plot showing the density of samples with varying degrees of minor variants in two different sampling periods (reddish March-September 2020 - n = 67 biologically independent samples and blueish October 2020 to January 2021 - n = 60 biologically independent samples). Data are presented as mean values (white dots into violin plots) +/- standard deviation (SD based on values between first and third quartile, black boxplot) and adjacent values (Upper and Lower) present with vertical black lines.
Supplementary information
Supplementary Tables 1–7 and Note.
Source data
Statistical source data used for Ct viral load comparison

Please Sign in (or Register) to view further.